PAT, PATTERN
NAME
PAT, PATTERN - define sequence pattern.
SYNOPSIS
PAT = name_11 name_12 ... name_1m / ... / name_k1 name_k2 ... name_kn
PAT TOL number_of_allowed_errors
PATTERN = name_11 name_12 ... name_1m / ... / name_k1 name_k2 ... name_kn
PATTERN TOLERANCE number_of_allowed_errors
DESCRIPTION
The command PATTERN (garlic version 1.4 or later) may be used to define a
sequence pattern. It is more general than the command SEQUENCE. Each pattern
is a set of name lists, separated by slashes (/). In the following example,
a simple pattern of three residues is defined:
PAT = ASP ASN / PHE TYR TRP / ARG LYS
The first residue may be matched by either ASP or ASN, the second residue
may be matched by PHE, TYR or TRP, while the third residue may be matched
by ARG or LYS.
The macromolecular structure may be searched for the given pattern, using
the command SELECT PATTERN (short form: SEL PAT). The commands RESTRICT
and ADD may be also used to search for the fiven pattern. For the pattern
defined above, the following fragments will be selected:
ASP PHE ARG
ASP PHE LYS
ASP TYR ARG
ASP TYR LYS
ASP TRP ARG
ASP TRP LYS
ASN PHE ARG
ASN PHE LYS
ASN TYR ARG
ASN TYR LYS
ASN TRP ARG
ASN TRP LYS
WILDCARDS
The command PATTERN may be combined with wildcards. The character * (asterisk)
may be placed at any position in the pattern string, though it does not make
much sense at the first and at the last position. Example:
PAT = ARG LYS HIS / * / ARG LYS HIS / * / ARG LYS HIS
PATTERN TOLERANCE
TOLERANCE (short form: TOL) is the only keyword which may be combined with
the command PATTERN. It may be used to define the pattern tolerance, i.e. the
maximal number of allowed errors which may be tolerated while searching for
the specified pattern.
DELETIONS AND INSERTIONS
It is not possible to handle deletions and insertions directly. However,
a number of different patterns may be incorporated into a simple script,
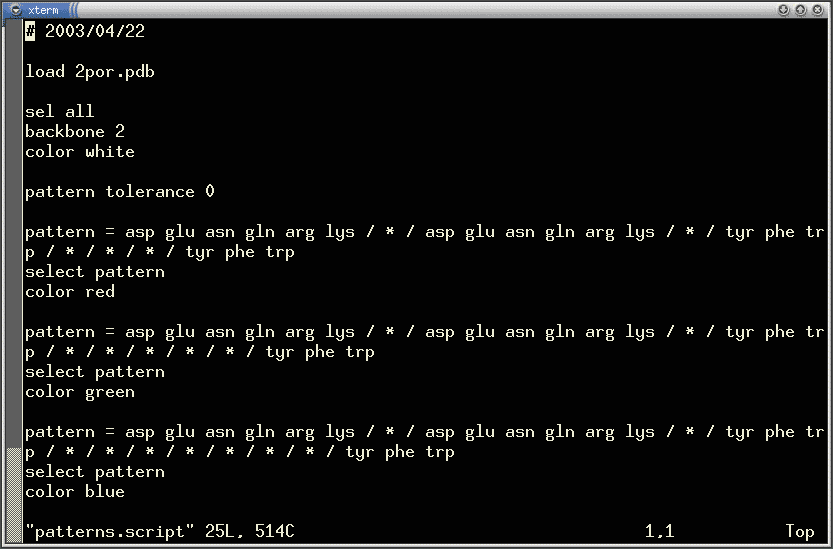
which may be used to handle deletions and insertions. The example below is
a screen shot of one such script (loaded into vi editor). The script contains
three patterns of different length. Each pattern was written as a single
line but it was wrapped by the editor because of length. Note that many
consecutive wildcards are used in this example.
NOTES
(1) The maximal number of residue names in each list is 30 (more than enough
for 20 standard residue names). The maximal number of lists in a pattern
is 100. If this is not enough, change MAX_PATT_LENGTH in defines.h file.
RELATED COMMANDS
The command SEQUENCE, combined with the keyword = (equality sign), may be
used to define a fixed sequence pattern. Of course, only one residue at
each position may be specified with the command SEQUENCE. The commands
SELECT, ADD and RESTRICT may be used to search the macromolecular structure
for the specified pattern.
