| SHORT FORM | LONG FORM |
|
WHE
WHE CLO WHE start_index end_index WHE start_index end_index CLO WHE OFF |
WHEEL
WHEEL CLOCKWISE WHEEL start_index end_index WHEEL start_index end_index CLOCKWISE WHEEL OFF |
| COMMAND | DESCRIPTION |
|
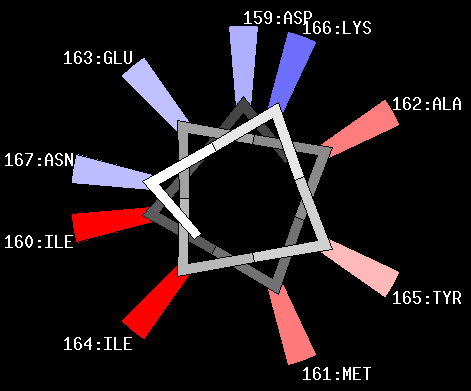
seq = asp ile met ala glu ile
scale eis angle 100 wheel |
Initialize the sequence fragment,
select Eisenberg hydrophobicity scale, set angle to 100 degrees and draw helical wheel. It is assumed that that the specified sequence fragment belongs to alpha helix. |
|
load 2por.pdb
seq from 1 scale eis angle 160 wheel 2-10 |
Load porin coordinates, copy the sequence
of this protein to the main sequence buffer, select Eisenberg hydrophobicity scale, set angle to 160 degrees and draw helical wheel for residues from 2 to 10. It is assumed that the specified sequence fragment belongs to beta strand. |
| whe clo |
Draw residues clockwise. This
gives the bottom view of a helix, which is more common in books. |
| whe off |
Return to default drawing mode.
Instead using this command, you can hit the ESCAPE key. |