 |
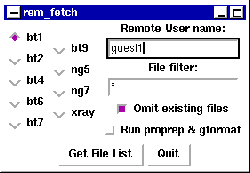
After data has been collected, it can be transferred using the UNIX command, rem_fetch. When this command is invoked, a graphic user interface (GUI) appears, as seen in Fig. 20. For the ``Remote User Name'' enter the directory used to collect data, for example, guest1 for [BT1.GUEST1] or sam.nano for [BT1.SAM.NANO]. Press the ``Get File List'' button to see a list of files displayed. By default, all files in the appropriate CNBS directory are then displayed (as seen in Fig. 21), except that files previously transferred into your current directory are not included. This behavior can be changed by deselecting the ``Omit existing files'' checkbutton, which causes all files to be displayed. It is also possible to select the files that will be listed using the ``File filter.'' For example, use of *ac* for a file filter restricts the file list to files containing the string ``ac''.
After the ``Get File List'' button is pressed, the window is modified to include the file list, as seen in Fig. 21. At this point it is possible to modify any of the previously described fields and then press the ``Get File List'' button to obtain an updated list.
Multiple files are selected by clicking on them while holding the shift key down, or groups of files can be selected by using control while clicking on them. Once a set of files has been selected, pressing the ``Transfer File(s)'' or the ``Transfer File(s) and Quit'' button causes the files to be transferred to the local computer. If the ``Run proprep & gformat'' checkbutton is selected, each file is individually processed by the proprep and gformat programs (see below) as the files are transferred.
Program proprep is used to convert .bt1 files into .raw files used by a number of programs, such as REFINE and are used as an intermediate for conversion to GSAS format. It can be used to add multiple runs, but this is better done in program gformat, if files for GSAS are to be produced.
The program can be used by typing:
proprep file
or
proprep file.bt1
Either command causes file file.bt1 to be read and a new file,
file.raw, to be created. Several (up to 50) files can be processed at
one time:
proprep file1 file2 file3
or even,
proprep abc*.bt1
where the .bt1 extension is assumed if not specified. The program
will produce an error message and fail if the .raw file exists.
It is important to look carefully at your data before execution of proprep. For instance, you may wish to exclude data below 5 degrees in detector 1, 10 degrees in detector 2, and 15 degrees in detector 3 using the option -cx.x (see below), as the background levels in these regions is often anomalously large. The default is to exclude data below 3.0, 8.0, and 13.0 degrees, respectively for detectors 1, 2 and 3, which would affect only data taken with the Ge(311) monochromator under standard data collection conditions.
A number of options can be supplied to proprep:
proprep [-s] [-k] [-1] [-cx.x] [-dx,y] [-p] file1 [file2] ...
or proprep can be used as a filter:
proprep -f [-k] [-1] [-cx.x] [-dx,y] < file.bt1 > file.raw
The full list of options accepted by proprep follows. Options can be combined
together where appropriate.
In the simplest form, program gformat is used by typing,
gformat abc.raw
to process abc.raw into abc.gsas. Note that
the .raw extension is assumed if not specified for input files, so that
the command
gformat abc
is equivalent to the previous command.
Up to 50 files can be specified on the command line. Each will be processed individually (unless -s is present, which causes the contents of the files to be averaged, see below). To process a series beginning with the letters ``abc'' you can use the commands
gformat abc*.raw
Background corrections are applied to each detector bank to give the best agreement for overlapping data. This correction can be omitted using the -o option. The program will not overwrite an existing .gsas file unless -c is specified.
When a zero correction is applied to each detector bank, data points will no longer ``line up'' with the data points in the output file. Thus, gformat will interpolate the intensity for each point when it bins. This creates some correlation between adjacent points, which is not strictly appropriate for correct least-squares statistics. If -m is used the points are placed into the closest bin without interpolation. This introduces a bit more scatter into the data, but is statistically more valid. The -m option causes your output step size to be double that of your input, which reduces the scatter that is introduced, so if you are going to use -m, you should probably collect data with a smaller step size 0.02 or 0.025 degrees).
The data for all detector banks is included in the .gsas file, unless the -L option is used (see below). A .inst file is created with the same file name as every .gsas output file. This contains the wavelength and other sundry information.
The .gsas and .inst files created by gformat can be e-mailed, ftp'ed etc. However, before using the files in GSAS, they need to be converted to direct access. This is done automatically for use with UNIX versions of GSAS by gformat. The direct access data files are named with all capital letters. When using e-mail or for non-UNIX versions of GSAS, transfer the lower-case named files.
A number of options can be supplied to gformat:
gformat [-s] [-c] [-o] [-m] [-Lxx] [-a] file1 [file2] ...
or gformat can be used as a filter:
gformat -f [-Lxx] [-o] [-m] [-a] < file.raw > file.gsas
The full list of options accepted by gformat follows. Options can be combined together as appropriate.
The -g option requires an absorption correction input file, abs.corr, which consists of a free-format file containing a one-line title followed by the number of Chebyshev terms (up to 30) and the Chebyshev polynomial coefficients. The polynomial is evaluated for
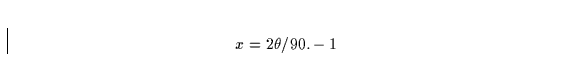
cmpr -readopt file1 file2 -readopt file3
The following read-options are available:
After data has been read, it can be plotted using the dialog box in Figure 23. Multiple files can be selected for plotting using shift plus the left mouse button and then using the ``Update Plot'' button (or double-clicking). It is also possible to change the way one or more files are displayed by selecting the files then selecting options for the Line, Color, or Symbol and then pressing ``Apply Changes.''
Note that when a file is plotted, it is possible to ``zoom-in'' using the left mouse button to click on two corners of the new region. The right mouse button zooms out and the middle mouse button opens a window for manual scaling.
The HKLGEN and EditCell dialog boxes (see Figure 24) are used
to generate the allowed reflection positions for a given set of unit cell
parameters and optionally using space group extinctions. In EditCell, these
reflection positions can be superimposed as vertical lines
on one or more sets of data. The
cell parameters can be changed and the reflection positions will shift as
the lattice constants change. Also, extinct reflections can be highlighted.
To see the indices for a reflection, shift-click on the line.
The cmpr program can also be used for peak fitting, as shown in Figure 25. The program uses the GPLSFT program, as written by David Cox and Larry Finger, to do the actual fitting.
To fit one or more peaks, first select the dataset to fit, then zoom-in to the region to fit using the left-mouse button and press ``Set range from graph'' (or manually enter the 2-theta range). Peaks may be entered or modified by clicking on the appropriate ``Set'' button and then by clicking on the appropriate location on the graph. The ``Use'' checkbuttons select if a peak is included in the fit. The remaining checkbuttons determine the variables that are refined when the ``Run GPLSFT'' button is pressed.
The cmpr program has other options. For example, the Rescale dialog can be used to multiply or add a constant to a dataset's x-axis or y-axis. It can also be used to change the units for plotting a dataset. For example, intensities can be shown on a logarithmic scale or 2-theta can be converted to 2-theta at different wavelength, or d-spaces or Q. PlotOptions controls how PostScript output is treated. A command for plotting the output, or the name to be used for saving output, can be specified here.
mount -v /a
the computer will respond:
/dev/fd0 on /a type vfat (rw,noexec,nosuid,nodev)
or will report an error.
(cd /a/; rem_fetch)
scp "guestbt1@jazz:sunysb/[a-z]*" /a/
to include subdirectories use scp -r ... in the above command.
ls /a/
umount -v /a
The computer will respond
/dev/fd0 umounted
If the computer responds
umount: /a: device is busy
you have a process running in the /a/ directory. Type
cd
in all UNIX windows or if need be log out or all windows (other
than the ICP process) on the console
until you can run the umount command without an error.