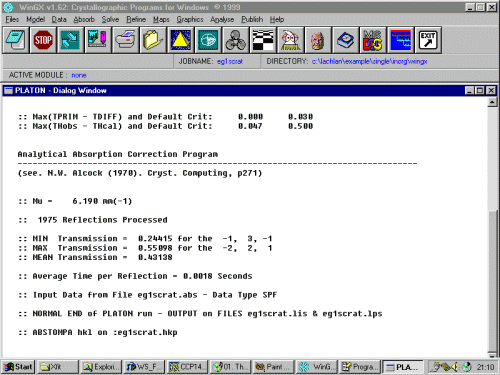
Click on Absorb, Numerical, Analytical menu option to enter the absorption correction option.
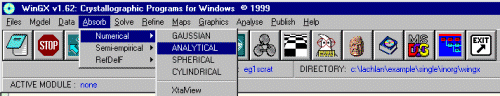
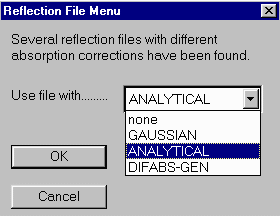
If WinGX detects an existing faces.def file, which has the indices and distance information, (implying you this is not the first absorption correction you have attempted on this sample) it will query whether you want to use this.
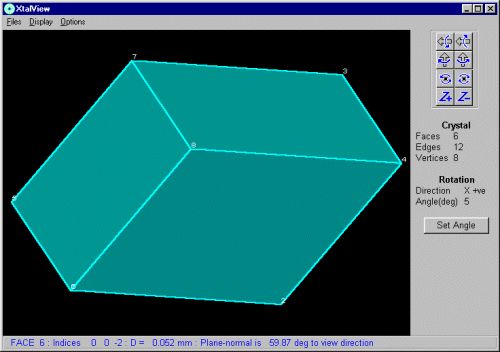
The faces.def file has the following format (distances in mm)
CELL 13.385 7.422 15.133 90.00 107.70 90.00 FACE 2 0 0 0.12000 FACE -2 0 0 0.12000 FACE 0 2 0 0.12000 FACE 0 -2 0 0.12000 FACE 0 0 2 0.05200 FACE 0 0 -2 0.05200
If you already have kosher a faces.def file to use, just used this, other wise refer to XtalView and Entering Faces for the First Time which will show you how to enter the faces via the WinGX GUI menu.