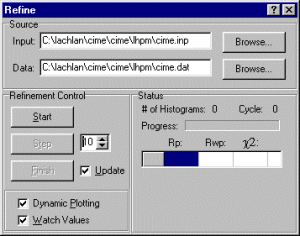
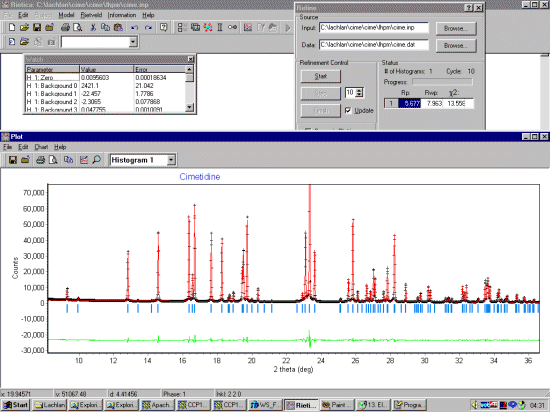
Setting up the Rietica-LHPM input file
Assuming you have the data file already, start Rietica and select File, New Input.
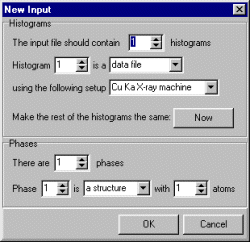
Select under "Phases" that you wish to do an extraction and unless you already have the synchrotron diffractometer defined as an "instrument", stick to the defaults and we will change these in the input file later.
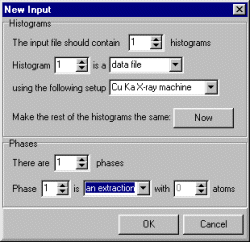
Select Model, General and setup the general information as shown in the following screen dump. Number of cycles, automatic Marquadt damping, data file format, we wish to obtain a EXTRA/Sirpow97 HKL file, and that we are using Synchrotron data.
This would be a good time to save and name the file. Select File, Save and browse to the directory that holds the Powder Diffraction data for cimetidine that you are using.
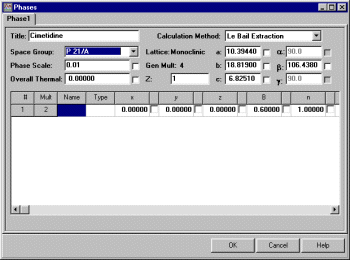
Select Model, Phases and input the basic information. That we want to do a Le Bail Extraction, Spacegroup and cell constants. the cell constants and two-theta offset should be from your unit-cell refinement so the Rietveld program should be right on the spot for starting the extraction. Le Bail fitting with an inaccurate cell can put you in a world of pain as the Le Bail fitting via the Rietveld software goes beserk.
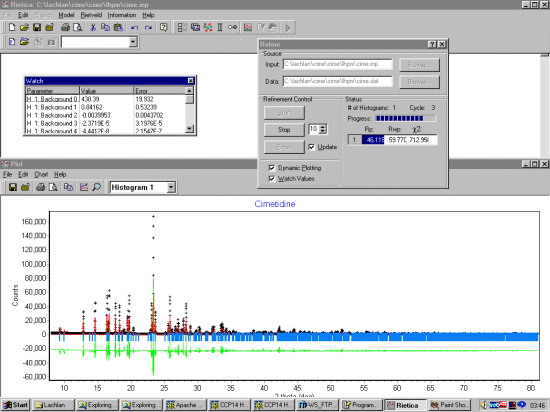
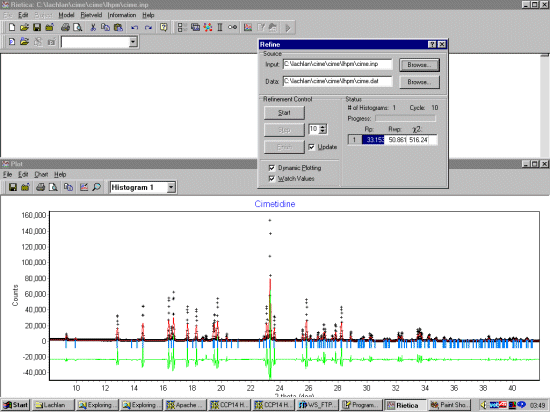
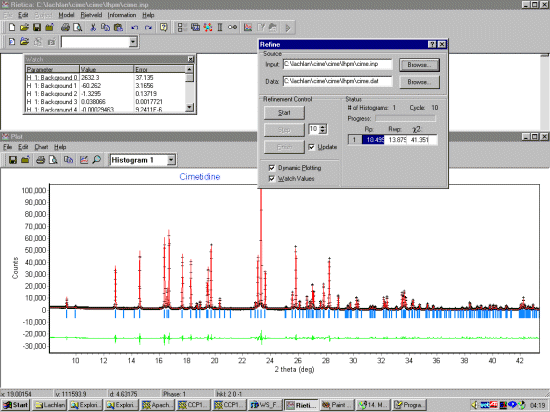
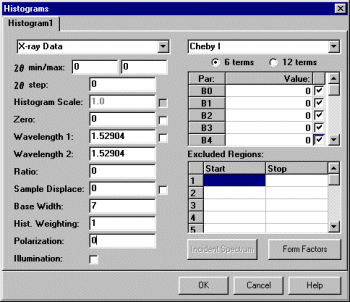
Select Model, Histograms and input the basic information. Setting the start, step and stop to zero tells LHPM to use the full pattern for the extraction. A Chebyshev function should be OK for fitting the background. Set the background function to refine.
Select Model, Sample and input the sample information. FJC Pseudo-Voight can be a flexible function to use that should work. As this is synchrotron data, set a thin width. Normally it would be good to fit a standard such as NIST/NBS Silicon or NIST/NBS Cerium Oxide to get good starting width and shape values. But if we don't have this information, starting off with a constant width should be OK.